Millions of people use the Python library Pandas to wrangle and analyze data. I, being one of those users, noticed a few months ago something peculiar: accessing rows by an index reference through .loc can be significantly slower when using double bracket notation [[]] than single bracket notation [], even when passing the same label. This is the story about how I ended up fixing a performance issue in the pandas source code because of this.
Contents
- The backstory: how would anyone ever notice this?
- Strange happenings: slow access for double brackets
- I’m not alone: finding GitHub issues filed by others
- Digging deeper: tracing my way through the source code
- The fix: binary search instead of re-indexing
- Wrap up
The backstory: how would anyone ever notice this?
Let’s say I’m analyzing the Auto MPG dataset from CMU, and I want to filter the dataframe by each of many car names. Intuitively, we might try indexing on the car name field and the using the pandas .loc method to filter our dataframe for each distinct car name:
1
2
3
4
5
6
import pandas as pd
# read Auto MPG data
df = pd.read_csv('https://raw.githubusercontent.com/mwaskom/seaborn-data/master/mpg.csv')
df.set_index('name', inplace=True) # set car name as the index
df.loc['plymouth duster']
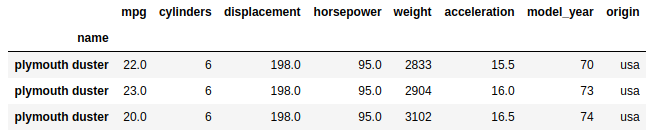
Great, but we’d have to be careful about using the right bracket notation with .loc though, as filtering for a name with only one instance will return a pandas Series object if there is only one match in the index. Roughly 80% of all car names have only one record in the table and the remaining 20% have multiple occurrences:
1
2
3
4
5
6
7
8
9
10
11
12
13
14
# make a histogram of number of records per name
name_counts = df.index.value_counts()
name_count_hist = name_counts.value_counts(normalize=True)
# plotly setup
import plotly.offline as py
py.init_notebook_mode(connected=True)
import plotly.graph_objs as go
# plot histogram
data = [go.Bar(x=name_count_hist.index, y=name_count_hist.values)]
layout = go.Layout(title='Name count distribution')
fig = go.Figure(data=data, layout=layout)
py.iplot(fig)
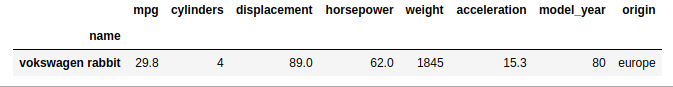
Choosing a name at random from the set of names with only one count, we can see that .loc with single bracket notation [] returns a different data structure if there is only one row in the index that matches the provided label:
df.loc['vokswagen rabbit'] # returns a series

If we wanted to consistently get a data frame back instead of a series sometimes and a frame other times, we’d have to use the .loc double bracket notation [[]]. More concretely,
(
type(df.loc['plymouth duster']),
type(df.loc['vokswagen rabbit']),
type(df.loc[['plymouth duster']]),
type(df.loc[['vokswagen rabbit']]),
)
returns (pandas.core.frame.DataFrame, pandas.core.series.Series, pandas.core.frame.DataFrame, pandas.core.frame.DataFrame). So we’d want to just use the double brackets notation [[]] across all car names to make sure we always get frames in return:
df.loc[['vokswagen rabbit']] # returns a dataframe, not a series
Strange happenings: slow access for double brackets
After settling on double brackets notation, I wasn’t super happy with the performance of my code. A quick comparison of double bracket notation vs single bracket showed double bracket notation isn’t much worse than single notation.
1
2
3
4
5
6
# artificially increase dataframe size to better measure asymptotic runtime
df_perf = pd.concat([df.iloc[:, 1]] * 10 ** 5)
# time performance of single vs double bracket notation
%timeit df_perf.loc['vokswagen rabbit'] # 460 ms
%timeit df_perf.loc[['vokswagen rabbit']] # 462 ms
But I knew that pandas .loc accesses were supposed to be faster if the index was sorted.
1
2
3
4
5
6
# sort data frame by index
df_perf.sort_index(inplace=True)
# time performance of single vs double bracket notation
%timeit df_perf.loc['vokswagen rabbit'] # 141 µs
%timeit df_perf.loc[['vokswagen rabbit']] # 406 ms
So this is interesting, and unexpected – it was significantly faster (~3000x difference) to access the same underlying data when using single bracket notation vs double bracket notation, but only if the data frame was sorted on index.
I’m not alone: finding GitHub issues filed by others
After a bit of digging, I found that I was not alone in noticing this issue. Pandas is open source and its source code is managed on GitHub, so I was able to find a filed issue for this exact problem. Here’s a minimal reproducible example from the issue, modified for readability and to account for changes in the API since filing (this issue was filed over three years ago):
1
2
3
4
5
6
7
8
9
10
11
12
import numpy as np
# create a dataframe of length 10^7 with a non-unique, monotonically increasing index
N = 10 ** 7
repeat_loc = 362
df_rand = pd.DataFrame(np.random.random((N, 1)))
df_rand.index = list(range(N))
df_rand.index = df_rand.index.insert(item=df_rand.index[repeat_loc], loc=repeat_loc)[:-1]
# benchmark performance
%timeit df_rand.loc[repeat_loc] # 225 µs
%timeit df_rand.loc[[repeat_loc]] # 633 ms
Digging deeper: tracing my way through the source code
I was curious about why performance was so much worse when using double brackets notation here, and decided to do some profiling with the python debugger pdb.
1
2
3
4
5
import pdb
# profile performance using pdb
pdb.set_trace()
df.loc[['vokswagen rabbit']]
In the screencast above, I traced my way through the .loc call stack until I saw something that sounded like it might be time consuming. The screencast ends at the _getitem_iterable method in pandas/core/indexing.py, where we see a call to labels._reindex_non_unique being made. This caught my eye because we just went out of our way to ensure that our data was indexed, and that our index was sorted for faster row accesses. So why does pandas re-index here if our data is already indexed?
Taking a step back, indexing makes it faster to access information later by copying and structuring information about the data in an “index” data structure; indexing saves lookup time at the cost of some upfront computation and storage. There are many ways of indexing (just look at the PostgreSQL documentation on indexes), but if we followed the call stack further we’d see here that our index is just an Int64Index object by printing its type in the pdb interface – according to the pandas documentation, this index is basically a sorted array.
If we trace a .loc call for single brackets with pdb, we’d eventually find ourselves in the core.indexes.base.get_loc method, where we reference a get_loc method in the self._engine object. In pdb, we can see that this object is of type pandas._libs.index.Int64Engine, which is implemented in Cython here. In the get_loc method there, we can see ultimately that the engine will locate values in the index through binary search (here and here).
So why reindex for every double brackets .loc call? The reindexing method we noticed in the pdb session above ends up referencing the same index engine, where it calls the get_indexer_non_unique method. Looking at the Cython source code, we can see pandas performs one pass through the entire index and stores a mapping from targets to index location. It then does a little bit of cleaning and restructuring, and returns the resulting index locations as the new (mini) index. This process runs in O(n + m) time where n is the length of the index and m is the number of targets. Accessing the rows from the index takes O(m) time after this, resulting in a total runtime complexity of O(n + m).
An alternative is to binary search, which pandas uses for a single brackets .loc call as we saw above. Binary search runs in O(log n) time for each of m targets or keys, resulting in a total runtime complexity of O(m log n). If m is large enough, then the repeated binary searches would be more costly than the linear scans from reindexing – as m approaches n then the rutime complexity approaches O(n log n) for repeated binary searches compared to only O(n) for reindexing. However if m were small enough relative to n, then a few binary searches would be faster than the full scans – as m approaches 1 then runtime complexity approaches O(log n) for repeated binary searches and O(n) for reindexing.
The fix: binary search instead of re-indexing
As someone who frequently benefits from open-source contributions, I decided it was worthwhile to contribute back to the community. Having identified the issue, I implemented a simple patch at the index engine level for the get_indexer_non_unique method to run repeated binary searches if the index is already sorted and there are only a few targets. I set the maximum number of targets to be 5, though a more principled approach should be taken in the future. I then opened a pull request which was accepted and merged into the master branch, and will be released in version 24.
Wrap up
The path from noticing a strange and unexpected behavior to merging a performance fix into production was quite enlightening (and rewarding) for me. I learned that the pdb debugger is useful for learning about not only your own code but others’ as well. I learned that indexing is far more complicated than I had initially imagined, but that there are clear(ish) wins to be made sometimes. Lastly, I learned that it feels good to contribute a fix to the community for an issue that had bothered me for some time.